Multi-Scale Representation Learning for Protein Fitness Prediction
Sep 26, 2024·,,,,,,,·
0 min read
Zuobai Zhang *
Pascal Notin *
Yining Huang
Aurélie Lozano
Vijil Chenthamarakshan
Debora Marks
Payel Das
Jian Tang
Abstract
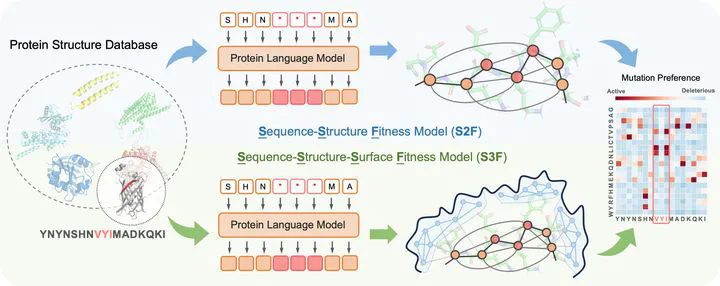
Designing novel functional proteins crucially depends on accurately modeling their fitness landscape. Given the limited availability of functional annotations from wet-lab experiments, previous methods have primarily relied on self-supervised models trained on vast, unlabeled protein sequence or structure datasets. While initial protein representation learning studies solely focused on either sequence or structural features, recent hybrid architectures have sought to merge these modalities to harness their respective strengths. However, these sequence-structure models have so far achieved only incremental improvements when compared to the leading sequence-only approaches, highlighting unresolved challenges effectively leveraging these modalities together. Moreover, the function of certain proteins is highly dependent on the granular aspects of their surface topology, which have been overlooked by prior models. To address these limitations, we introduce the Sequence-Structure-Surface Fitness (S3F) model — a novel multimodal representation learning framework that integrates protein features across several scales. Our approach combines sequence representations from a protein language model with Geometric Vector Perceptron networks encoding protein backbone and detailed surface topology. The proposed method achieves state-of-the-art fitness prediction on the ProteinGym benchmark encompassing 217 substitution deep mutational scanning assays, and provides insights into the determinants of protein function. Our code is at https://github.com/DeepGraphLearning/S3F.
Type
Publication
In The 38th Annual Conference on Neural Information Processing Systems (NeurIPS 2024)